Building a Webapp for Species Geofencing
One of the issues we experienced with speciesnet classification was results where the species was not valid for our location. In speciesnet - specifying a geofence coordinate will cause missclassifications to move up to the highest level hierarchy - in most cases, this classifies an animal as “animal.”
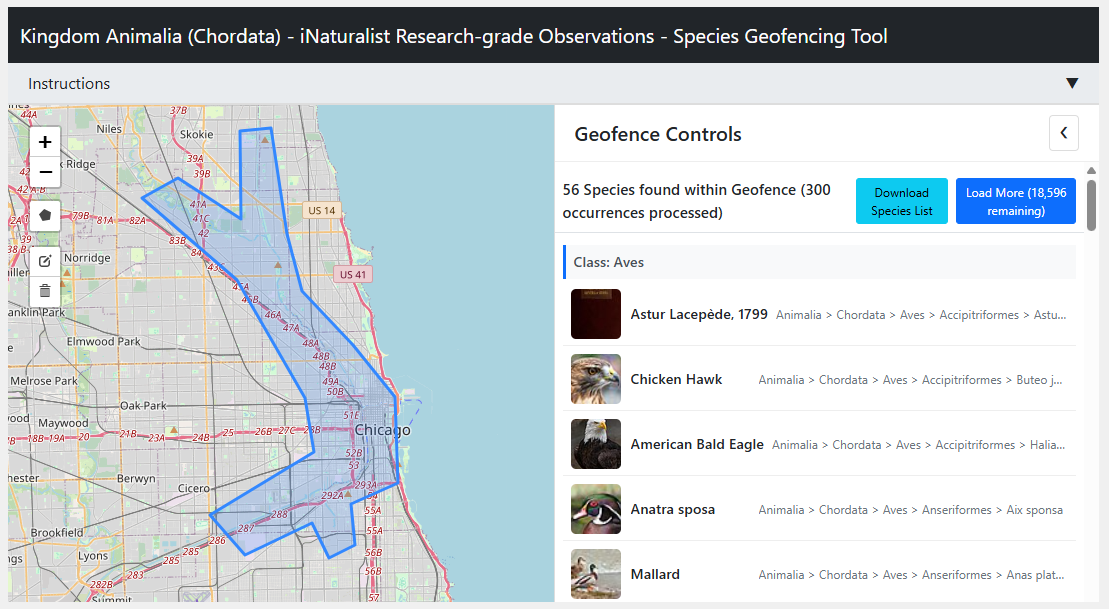
So, to help validate lists of species relevant to our area, I built a webapp for looking up species.
A well known issue
I ran a test of google’s species net and wrote about the results here.
Species geofencing is problematic. Animals don’t seem to care where you draw lines on a map and they don’t have “guest books” where they can sign their names. We rely on human observation to tell us if a species is found in an area or not.
Of all the public resources for these observations - the Global Biodiversity Information Facility (GBIF) API has a great search query representation for polygons. This search is baked into their tools, but I wanted to be able to filter the results for unique species instead of by occurence.
The ability to draw on a map and be presented with a unique list of confirmed species was the main goal.
Cursor, Jules, and GitHub CoPilot created the majority of the code - we started in Flask and then I asked for a refactor for a full front end site for easy hosting (wow).
The project repo and branch can be found on my GitHub.
Cleaning AI computer vision results based on polygon geofence occurrences
This polygon was used to then submit a query to the GBIF dataset saved on Google BigQuery.
WITH unique_species AS (
SELECT DISTINCT
class,
`order`,
family,
genus,
species,
taxonkey
FROM
`bigquery-public-data.gbif.occurrences`
WHERE
ST_WITHIN(
ST_GEOGPOINT(decimallongitude, decimallatitude),
ST_GEOGFROMTEXT('POLYGON((
-87.69081115722656 42.005312912238956,
-87.66952514648438 41.955818412264705,
-87.61596679687501 41.905774595463853,
-87.60910034179689 41.85779952612765,
-87.62626647949219 41.815801430687642,
-87.7196502685547 41.808127409160392,
-87.71690368652345 41.842908943268263,
-87.67982482910158 41.88533726561532,
-87.72377014160158 41.946119107705776,
-87.78625488281251 41.99051961904691,
-87.69081115722656 42.005312912238956
))')
)
AND LOWER(phylum) = "chordata" # This is the phylum that includes mammals and birds
LIMIT 1000
)
SELECT
t1.*,
ARRAY_AGG(DISTINCT countrycode IGNORE NULLS) AS country_codes
FROM unique_species t1
LEFT JOIN `bigquery-public-data.gbif.occurrences`
USING(taxonkey)
GROUP BY
class,
`order`,
family,
genus,
species,
taxonkey
Less than 1000 were returned in this query, but that could be a large bill if the polygon area were huge
Using geofencing for validated species lists vs. speciesnet
There are 3 datasets worth using to clean up missclassified speciesnet data:
- The predictions dataset itself ( from our non-geofenced speciesnet inference on our images )
- The published SpeciesNet taxonomy_release.txt file ( a list of all possible output classifications )
- A geofenced dataset from the polygon created using the webapp ( filtering gbif occurrences by location )
Each dataset has its own structure, but ultimately they share a listing of the class/order/family/genus/species heirarchy of taxonomic classification.
Joining and filtering to find what species should be valid for a location is possible almost entirely in python-pandas.
The notebook for this analysis can be found here.
Example of matching vs non-matching lists:
| matching (speciesnet&gbif-chi) | non_matching (speciesnet!gbif-chi) |
|----------------------------------|---------------------------------------|
| blank | human |
| bird | western pond turtle |
| mallard | anseriformes order |
| american coot | domestic dog |
| northern raccoon | reptile |
| great blue heron | domestic cattle |
| vehicle | wild turkey |
| eastern cottontail | central american agouti |
| brown rat | white-tailed deer |
| domestic cat | nutria |
| muskrat | crocodile |
| wood duck | wild boar |
| coyote | mammal |
| canada goose | common tapeti |
| american beaver | tome's spiny rat |
| eastern gray squirrel | ocellated turkey |
| domestic horse | rodent |
| american robin | collared peccary |
| white-crowned sparrow | spotted paca |
| sylvilagus species | madagascar crested ibis |
| song sparrow | canis species |
| snowy egret | red squirrel |
| california quail | bushy-tailed woodrat |
| north american river otter | pronghorn |
| eastern chipmunk | eastern red forest rat |
| <NA> | domestic chicken |
| <NA> | blood pheasant |
| <NA> | white-lipped peccary |
| <NA> | red acouchi |
| <NA> | desert cottontail |
| <NA> | plains zebra |
| <NA> | rufescent tiger-heron |
| <NA> | owl |
| <NA> | common wombat |
| <NA> | bearded pig |
| <NA> | fossa |
| <NA> | nine-banded armadillo |
| <NA> | guenther's dik-dik |
Officially not in Illinois at all:
| common_name | state_code | |--------------------------|------------------------------------------------------| | bushy-tailed woodrat | [CO, CA, WY, OR, UT, WA, NM, MT, SD, NV, ID, ...] | | central american agouti | [TX] | | common wombat | [NM, WA] | | crocodile | [FL] | | desert cottontail | [CA, AZ, CO, NM, TX, NV, UT, WY, MT, NE, SD, KS, ...] | | fossa | [NE, TX] | | guenther's dik-dik | [TX] | | ocellated turkey | [CA, FL, HI, OK] | | plains zebra | [CA, NM, TX, WA, AK, CO, MA, OH, OR, UT] | | pronghorn | [WY, CO, NM, UT, AZ, SD, MT, OR, ID, TX, NV, CO, ...] | | red acouchi | [NE, NY, WA] | | rufescent tiger-heron | [HI] | | spotted paca | [CO, CA, RI, UT, WA] | | western pond turtle | [CA, OR, NV] | | white-lipped peccary | [TX, CO, TX] |
Note we didn’t apply any geofencing to this run of speciesnet - so it’s expected to return the highest confidence match without ‘rolling-up’ to the higher taxonomy level.